DNA Replication/PCR in bacteria and Gene Transfer in Bacteria
Introduction
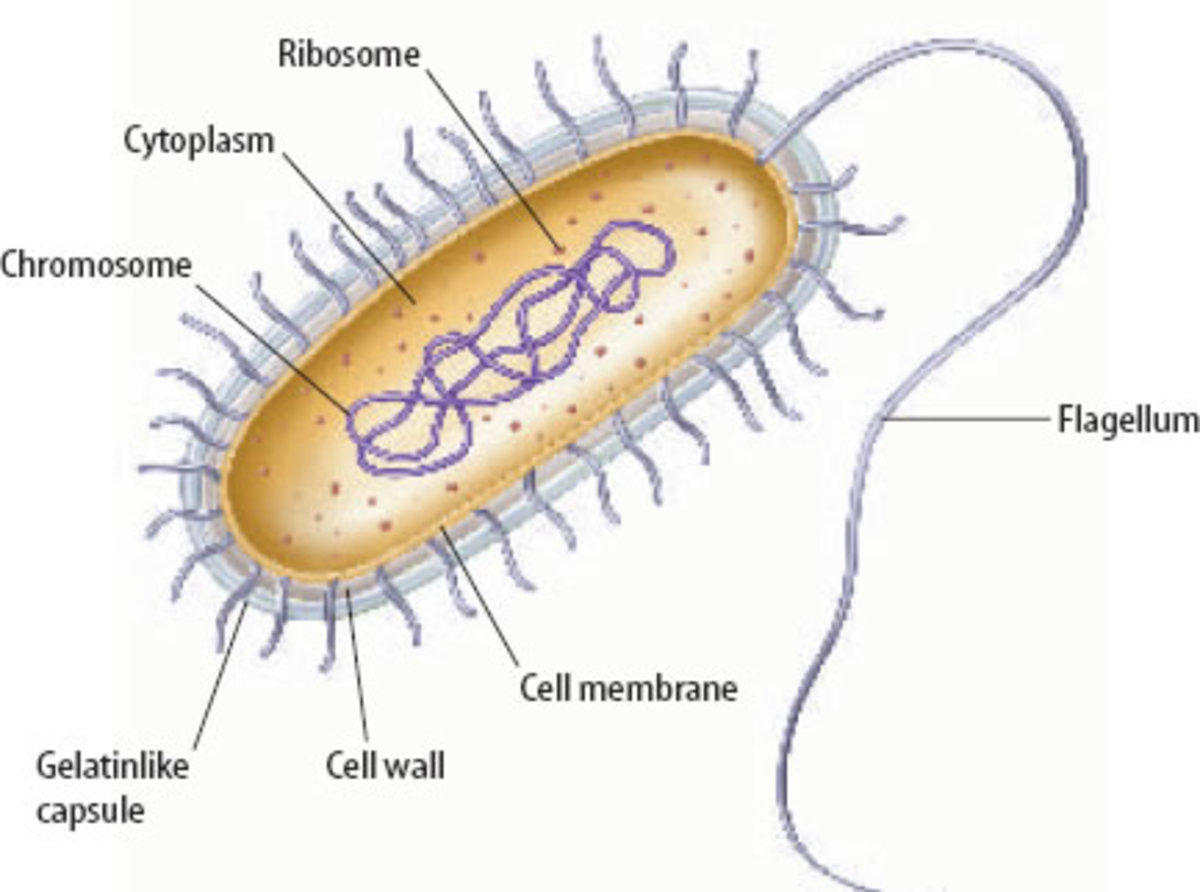
To sustain a bacterial population, it is imperative that cells divide, however, before the physical split of a daughter cell; it is important that the DNA has been copied accurately. The study of the cell-cycle steps in bacteria is typically divided into three phases: the time between cell division (cell birth) and the beginning of the chromosome duplication. The period required to complete DNA replication (the lengthening of DNA). The last phase, which goes from the conclusion of DNA replication until the end of cell division. In the best-growing conditions, DNA replication starts instantaneously after cell division in most cells. Since the replication of the chromosome consumes more time than necessary for cell division under the most favorable culture conditions.
Stages of DNA replication per cell cycle
Initiation of the DNA replication
In bacteria, the course of DNA replication begins in a particular DNA section called “origin of replication” (ori) where multi-protein complexes are situated. It adds additional initiator proteins to form the Pre-Replicative complex (pre-RC) whose principal function is to facilitate the aperture of duplex DNA to allow the loading of the replicative DNA helicase. The action of this DNA helicase assists the entry and assembly of a large multi-subunit molecular apparatus, the replisome.
DNA-A is the main protein required to assemble the pre-RC
The significant step for the successful duplication of DNA is the unfolding of the DNA strands at the oriC region, and this is assisted by the orisome (proteins-oriC complex). The complex mainly comprises of the action of the initiator protein DnaA. It is believed that the DnaA-oriC complex forms a circular pentamer, which is stabilized by interactions between the DnaA units. The development of these complexes initiates the unwinding of DNA strands on the commencement of replication. The domain IV of Dna-A has a helix-turn-helix pattern that permit it to interact with the DnaA-box of oriC.
Lengthening of DNA
The single strands of DNA (ssDNA) are stabilized by a protein known as single-stranded DNA binding protein (SSB). SSB binds to single DNA-strands in a tetramer form through its N-terminal domain, which touches the DNA.
Termination of DNA replication
The end of DNA replication occurs place when the replisome helicase DnaB6 on the leading strands meets with a protein known as Tus. Tus recognizes and is attached to sites for termination of DNA replication (ter). The sites are physically set in positions opposite to the oriC. During the collision of Tus with the helicase, a trap is developed that prevents further advancement of the replicative apparatus in the leading strand and remains in place until the replicative machinery on the lagging strand arrives at this position.
PCR technique
The mechanism and agarose gel electrophoresis
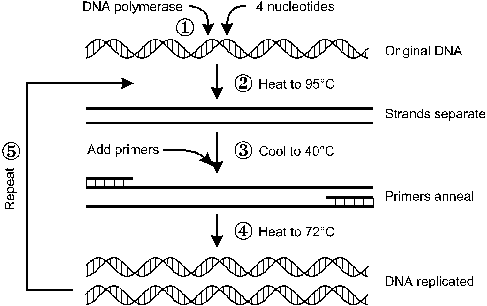
Polymerase chain reaction is considered a scientific method applied in molecular biology to amplify a portion of DNA across some magnitudes thus generating millions of copies of a particular DNA sequence. It can be thought of as a molecular photocopier.
The basic principle of PCR is simple: One DNA molecule is used to make two copies, then four, then eight. Specific proteins called polymerases facilitate this continuous doubling. These are enzymes that can join single DNA building blocks to form long molecular threads. Polymerases require DNA components, which are the nucleotides that consist of the four bases; adenine (A), cytosine (C) thymine (T), and guanine (G).
Agarose gel electrophoresis is considered a robust method of separation that is frequently used to analyzeDNA fragments that are generated by enzyme restriction. It is also a convenient method of analysis used for determining the size of DNA molecules that range from 500 to 30,000 base pairs. It can also be applied to the separation of other charged biomolecules such as RNA, proteins, and dyes. The separation medium is a gel that is obtained from agarose (a polysaccharide derivative of agar). It is obtained from the same seaweed as bacterial agar that is used in microbiology, including a food product called agar, which is applied in the preparation of a gelatin-like dessert in Asian cuisine. Agarose is a non-toxic substance because it comes from the same source as the food product agar. However, the gel does contain a buffer for conductivity.
PCR Technology
Gene transfer
Gene transfer is the transfer of a gene from one DNA molecule to another DNA molecule. Gene transfer does represent a new possibility for treatment of certain rare genetic disorders and common diseases that are multifactorial by changing the expression of a person's genes.
Gene transfer may be targeted to somatic or germ (sperm and egg) cells. In the somatic gene transfer, the genome of the recipient is altered, but the change does not get moved to the subsequent generation. In germline gene transfer, the parents' sperm cells and egg are modified with the aim of passing on these changes to their offspring. Germline gene transfer is currently not actively being studied, at least in larger humans and animals.
Conjugation in bacteria
Mechanism of conjugation
Bacterial conjugation is termed as the transfer of DNA information from a living bacterium (donor) to another bacterium (recipient). Plasmids are considered as small autonomously replicating circular pieces of circular double-stranded DNA. Conjugation involves the transfer of plasmids from a donor bacterium to recipient bacterium (Dale and Park 101). Transfer of plasmids in Gram-negative bacteria occurs only between strains of the same species or species that are closely related. Cells that possess F-plasmids are F+ (male) and always act as donors while those that lack this type of plasmid are termed F- (female) and always act as the recipient. Every Gram-negative F+ bacterium contains 1 to 3 sex pili, which bind to a particular outer membrane protein on recipient bacteria to trigger mating. After that, the sex pilus retracts thus bringing the two bacteria into contact with each other and the two cells get bound at a point of direct envelope-to-envelope contact.
In bacteria that is Gram-positive, sticky surface molecules get produced finally bringing the two bacteria into contact with one another. Donor bacteria that are Gram-positive do synthesize adhesins, which make them aggregate with the recipient cells, but sex pili do not get involved. DNA then gets transferred from the donor to the recipient.
The Transformation in bacteria
The mechanism of transduction
Transduction happens when there is a phage transfer bacterial DNA from one bacterium (donor) to the other (recipient). Occasionally, viral phages insert bacterial chromosomal DNA into their head instead of viral DNA. When this phage infects a bacterium that is defective for the A-gene (a-). The phage injects its DNA in the host.
When a phage attacks a bacterial cell, it inserts its DNA into the cell. The viral DNA is replicated severally, and viral the genes are expressed, producing the proteins that make up the viral capsid and nucleases that cleave the host genome into fragments. The newly copied viral DNA molecules are stored into viral capsids, and the bacterial cell is lysed, thus releasing hundreds of viral progeny, these then go on to infect other cells.
Transposition in bacteria
The mechanism of transposition
Transposition is a DNA recombination reaction that results in the translocation of a discrete DNA segment called a transposable element or transposon from a donor site to one of many nonhomologous target sites. Transposition can also promote other kinds of DNA rearrangements including deletions, inversions, and replicon fusions. Such rearrangements of DNA can have profound effects. It can result in the stable acquisition of new genetic information, for example, the perfection of viral DNA into a host chromosome.
At the heart of all transposition reactions is the insertion of a discrete DNA segment (the transposon) into a nonhomologous target site. Transposons insert such that the newly added element is flanked by short direct repeats of a sequence that existed only once in the target DNA. Since the element translocates between many different non-homologous sites, these duplications are not constant in sequence.
The detailed structures of trans- position products can vary considerably, depending on the relative locations of substrate transposon ends and the target site to which they will join, and on the particular type of transposition reaction.
Gene mapping (The mechanism of gene mapping)
"Gene mapping" is the mapping of genes to particular locations on chromosomes. It is a critical step in the understanding of genetic diseases. There are two forms of gene mapping: Genetic Mapping - using linkage analysis to determine the relative location between a pair of genes on a chromosome and Physical Mapping - using all available methods or information to determine the final position of a gene on a chromosome. The ultimate goal of gene mapping is to clone genes, especially disease genes. When a gene is cloned, the gene's DNA sequence and study of its protein product can be determined.